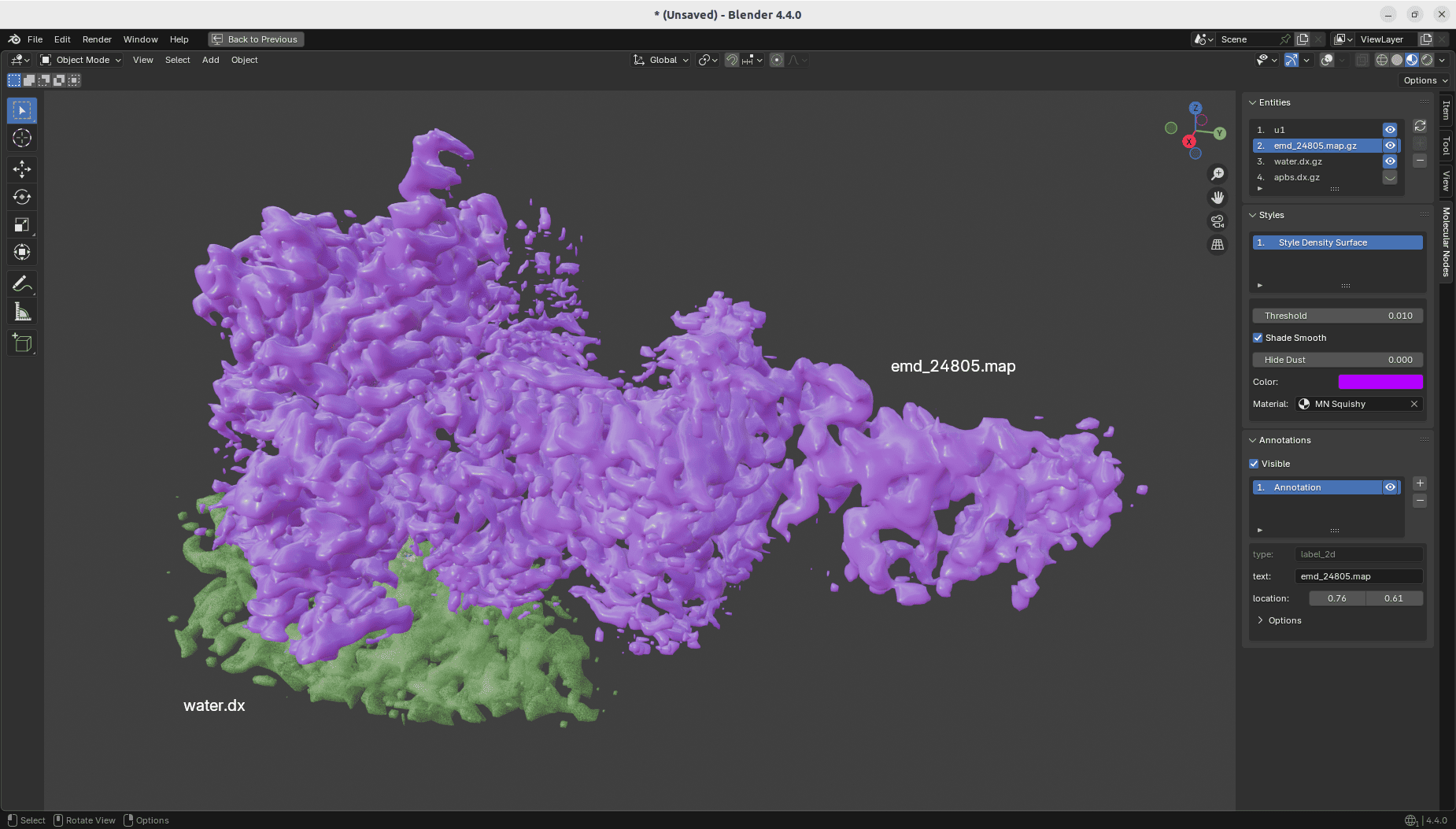
GSoC 2025 - Week 9
This week, I got started on the density component visualization. I initiated discussion #914 outlining the approaches and started implementing them.
I made the following progress:
- PR #916 (open)
- Fixed the density entity to work with the style interface (for both API and GUI)
- Added GUI support for density entities to show up in the 3D viewport panel
- Added annotations support for density entities as well
- Fixed a few other bugs encountered during this process
- PR #917 (open)
- Refactored density entity to use MDAnalysis’s GridDataFormats library instead of
mrcfile.GridDataFormatssupportsMRCfiles as well - Added a new Grids class that supports all the grid formats of the
GridDataFormatslibrary.dx,.plt,.ccp4,.mrc,.map,.pickle, (and compressed variants like.dx.gz,.ccp4.gz,.ccp4.bz2,.mrc.gz,.mrc.bz2,.map.gz,.map.bz2)
- Added support for different grid data types based on what
openvdbsupports - Support both older
pyopenvdb(< blender 4.4.0) and neweropenvdblibrary for generating and loading.vdbfiles in Blender - Min, Max support for thresholds in node socket inputs based on grid values
- Refactored density entity to use MDAnalysis’s GridDataFormats library instead of
Here are some learning from this week:
- Blender’s
openvdbsupport usesnanogridto check and warn for leaks. Anyvdbgrids created must be ensured they are cleaned up to avoid memory errors, especially inpytests - Blender seems to have some wierd caching of contents loaded from
vdbfiles. Overwriting avdbfile that is already loaded and re-loading it as a new object will update the old object as well - Though
GridDataFormatssupports a lot of commonly used grids in molecular simulations, most of them are limited to orthorhombic unitcells
Next week, I will fix any issues that come up during the review of the open PRs and wrap up work on densities by adding a new style node for ISO surfaces that allows different colors for positive and negative ISO values and also slicing along all the major axes.
This post is licensed under CC BY 4.0 by the author.